MACSE
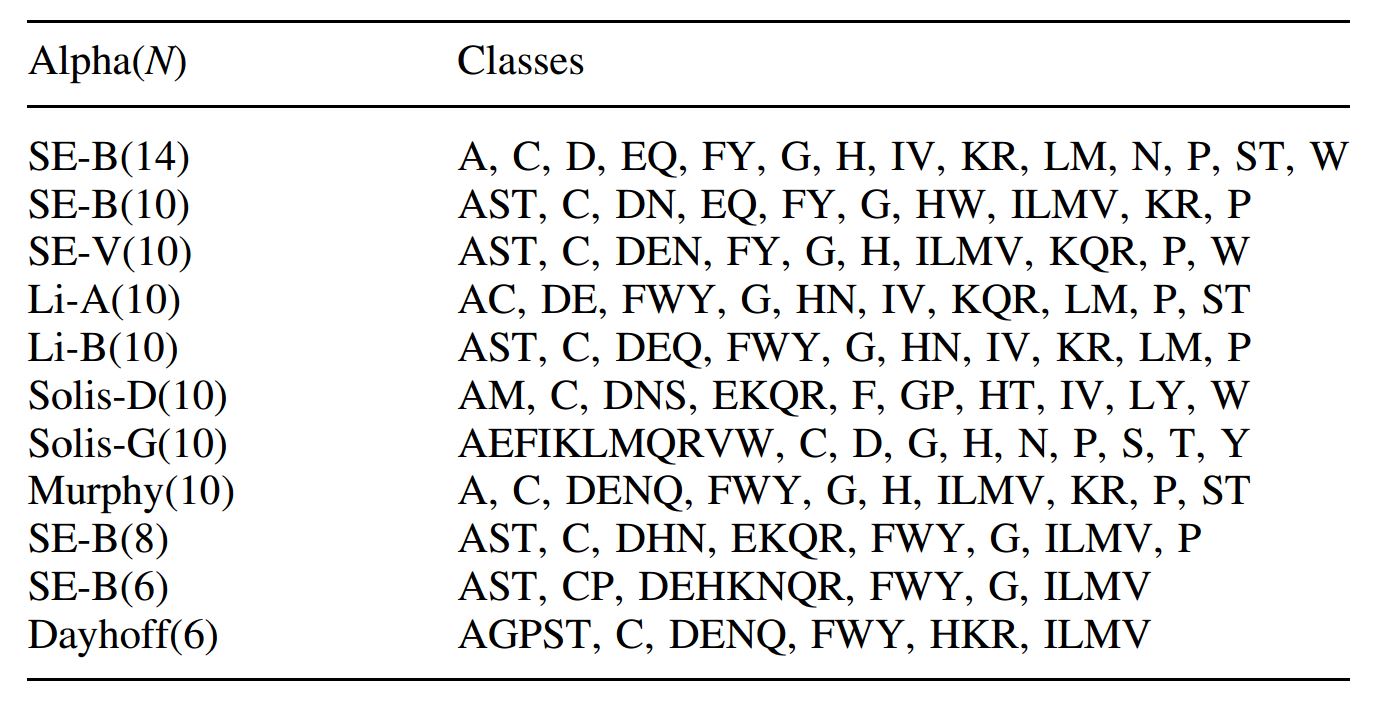
Compressed amino acid alphabet
Folder: samples/delegations/alphabet/
In some MACSE subprograms, you can use the alphabet_AA option to change the default compressed amino acid alphabet used by MACSE to compute initial pairwise distances or to identify homologous fragments. For instance the command to select the Dayhoff_6 alphabet is:
- java -jar macse.jar -prog translateNT2AA -seq sequences.fasta -use_compressed_alphabet_ON -alphabet_AA Dayhoff_6
The proposed alphabets are those listed in the Edgar 2004 paper.