Alternative Splicing
Alternative Splicing
Domestication is known to strongly reduce genomic diversity through population bottlenecks.
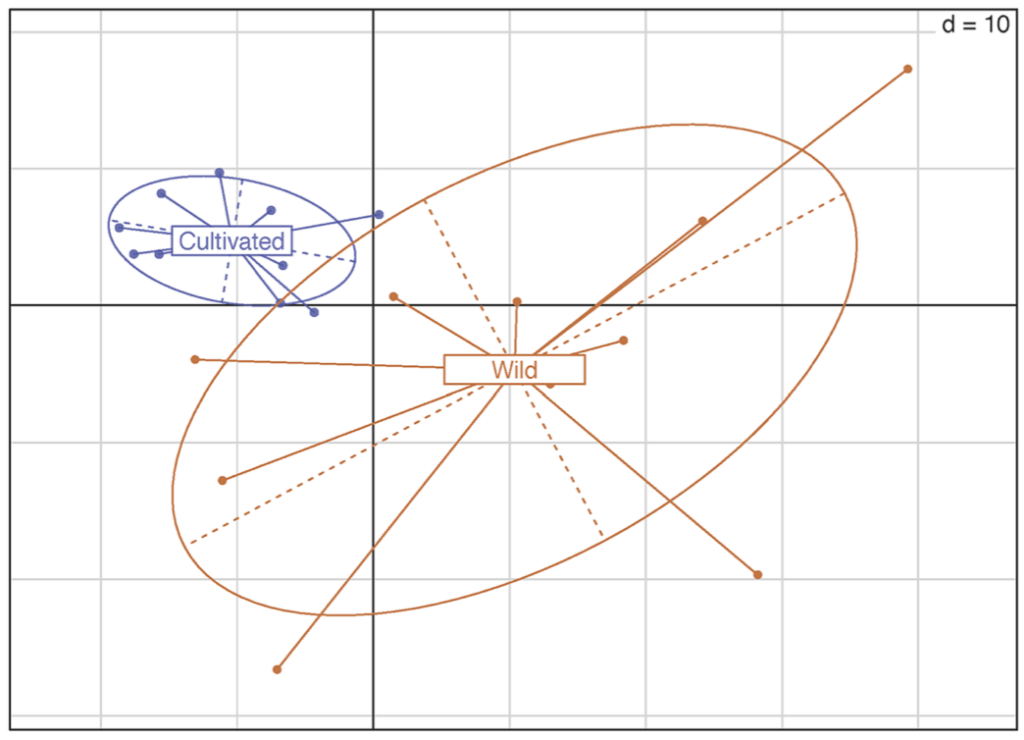
The resulting loss of polymorphism has been thoroughly documented in numerous cultivated species. Here we investigate the impact of domestication on the diversity of alternative transcript expressions using RNAseq data obtained on cultivated and wild sorghum accessions (ten accessions for each pool). In that aim, we focus on genes expressing two isoforms in sorghum and estimate the ratio between expression levels of those isoforms in each accession.
Noticeably, for a given gene, one isoform can either be overexpressed or underexpressed in some wild accessions, whereas in the cultivated accessions, the balance between the two isoforms of the same gene appears to be much more homogenous. Indeed, we observe in sorghum significantly more variation in isoform expression balance among wild accessions than among domesticated accessions. The possibility exists that the loss of nucleotide diversity due to domestication could affect regulatory elements, controlling transcription or degradation of these isoforms. Impact on the isoform expression balance is discussed. As far as we know, this is the first time that the impact of domestication on transcript isoform balance has been studied at the genomic scale.
This could pave the way towards the identification of key domestication genes with finely tuned isoform expressions in domesticated accessions while being highly variable in their wild relatives.